New device can diagnose Covid-19 from saliva samples | MIT News
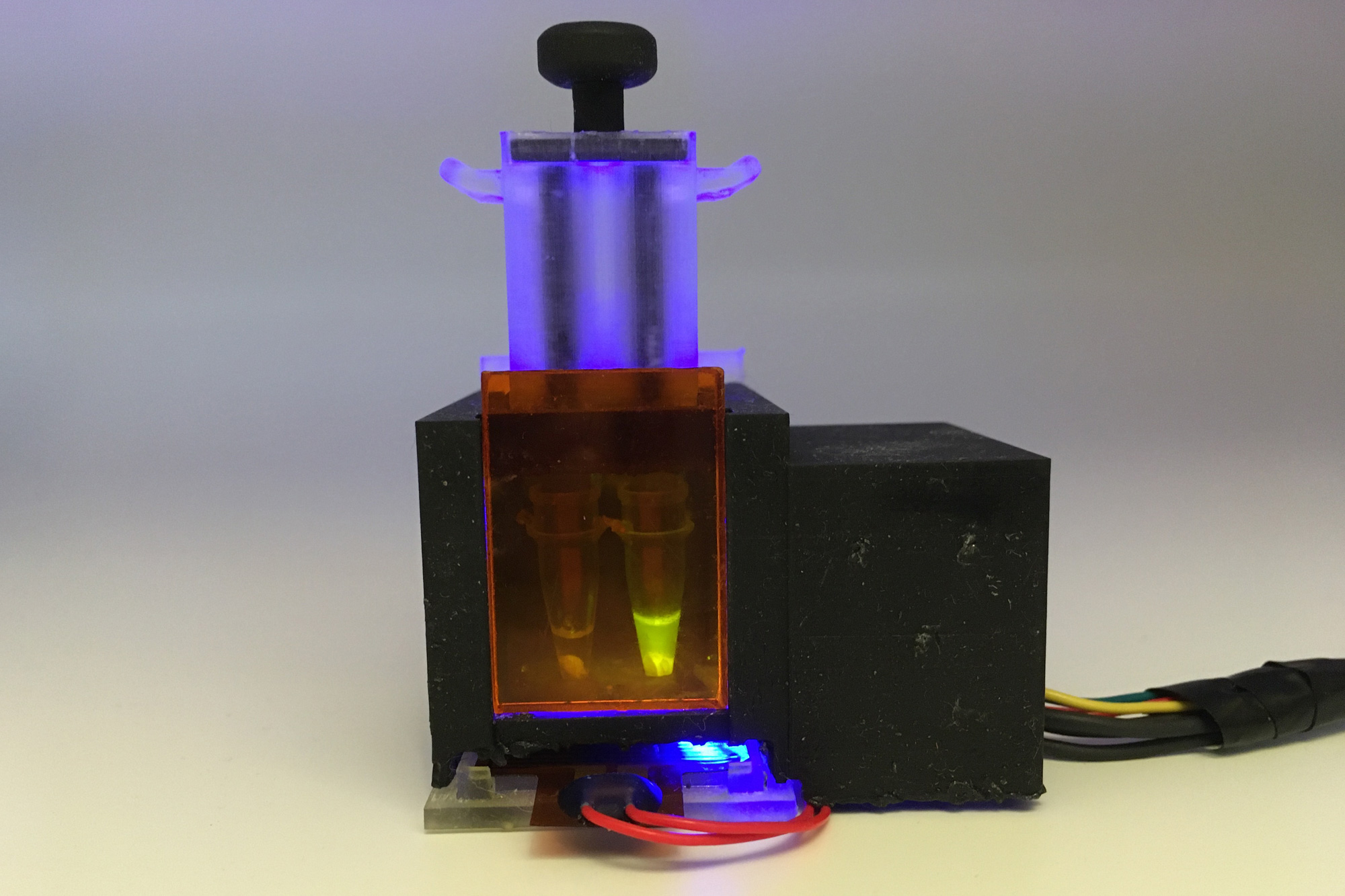
Engineers at MIT and Harvard College have designed a compact tabletop machine that can detect SARS-CoV-2 from a saliva sample in about an hour. In a new analyze, they showed that the diagnostic is just as precise as the PCR exams now applied.
The system can also be made use of to detect distinct viral mutations joined to some of the SARS-CoV-2 variants that are now circulating. This consequence can also be acquired in just an hour, perhaps building it considerably much easier to keep track of various variants of the virus, specifically in areas that never have entry to genetic sequencing facilities.
“We shown that our platform can be programmed to detect new variants that emerge, and that we could repurpose it very quickly,” suggests James Collins, the Termeer Professor of Health-related Engineering and Science in MIT’s Institute for Health care Engineering and Science (IMES) and Office of Biological Engineering. “In this examine, we targeted the U.K., South African, and Brazilian variants, but you could easily adapt the diagnostic platform to handle the Delta variant and other types that are emerging.”
The new diagnostic, which relies on CRISPR engineering, can be assembled for about $15, but people prices could appear down drastically if the devices were being developed at substantial scale, the researchers say.
Collins, who is also a member of the Wide Institute of Harvard and MIT, is the senior writer of the new study, which appears today in Science Innovations. The paper’s lead authors are Helena de Puig, a postdoc at Harvard University’s Wyss Institute for Biologically Influenced Engineering Rose Lee, an instructor in pediatrics at Boston Children’s Hospital and Beth Israel Deaconess Clinical Centre and a viewing fellow at the Wyss Institute Devora Najjar, a graduate college student in MIT’s Media Lab and Xiao Tan, a medical fellow at the Wyss Institute and an teacher in gastroenterology at Massachusetts Standard Hospital.
A self-contained diagnostic
The new diagnostic is primarily based on SHERLOCK, a CRISPR-based device that Collins and some others initial claimed in 2017. Components of the technique contain an RNA information strand that allows detection of specific target RNA sequences, and Cas enzymes that cleave people sequences and produce a fluorescent signal. All of these molecular elements can be freeze-dried for very long-expression storage and reactivated on publicity to h2o.
Very last calendar year, Collins’ lab commenced doing the job on adapting this technology to detect the SARS-CoV-2 virus, hoping that they could layout a diagnostic product that could produce immediate effects and be operated with minimal or no experience. They also desired it to operate with saliva samples, earning it even easier for consumers.
To attain that, the researchers experienced to integrate a essential pre-processing action that disables enzymes named salivary nucleases, which demolish nucleic acids this kind of as RNA. As soon as the sample goes into the gadget, the nucleases are inactivated by warmth and two chemical reagents. Then, viral RNA is extracted and concentrated by passing the saliva by a membrane.
“That membrane was critical to gathering the nucleic acids and concentrating them so that we can get the sensitivity that we are exhibiting with this diagnostic,” Lee states.
This RNA sample is then uncovered to freeze-dried CRISPR/Cas factors, which are activated by automatic puncturing of sealed water packets inside the unit. The just one-pot response amplifies the RNA sample and then detects the target RNA sequence, if present.
“Our target was to make an solely self-contained diagnostic that needs no other machines,” Tan says. “Essentially the patient spits into this product, and then you push down a plunger and you get an answer an hour afterwards.”
The researchers intended the gadget, which they contact minimally instrumented SHERLOCK (miSHERLOCK), so that it can have up to four modules that each individual appear for a unique target RNA sequence. The primary module is made up of RNA guidebook strands that detect any pressure of SARS-CoV-2. Other modules are unique to mutations affiliated with some of the variants that have arisen in the past calendar year, which include B.1.1.7, P.1, and B.1.351.
The Delta variant was not yet prevalent when the researchers done this examine, but because the technique is previously developed, they say it must be clear-cut to style and design a new module to detect that variant. The process could also be effortlessly programmed to monitor for new mutations that could make the virus additional infectious.
“If you want to do additional of a broad epidemiological study, you can layout assays in advance of a mutation of issue appears in a population, to check for possibly hazardous mutations in the spike protein,” Najjar states.
Monitoring variants
The researchers very first tested their machine with human saliva spiked with artificial SARS-CoV-2 RNA sequences, and then with about 50 samples from sufferers who had analyzed beneficial for the virus. They observed that the system was just as precise as the gold common PCR exams now made use of, which call for nasal swabs and take additional time and noticeably extra components and sample managing to produce final results.
The unit creates a fluorescent readout that can be found with the naked eye, and the scientists also designed a smartphone application that can study the benefits and deliver them to general public well being departments for less difficult monitoring.
The scientists believe that their system could be generated at a expense as minimal as $2 to $3 for every product. If approved by the Fda and created at significant scale, they visualize that this type of diagnostic could be beneficial possibly for individuals who want to be capable to exam at house, or in health care centers in areas with out common obtain to PCR screening or genetic sequencing of SARS-CoV-2 variants.
“The capacity to detect and observe these variants is important to productive community wellbeing, but unfortunately, variants are at this time identified only by nucleic acid sequencing at specialised epidemiological facilities that are scarce even in source-loaded nations,” de Puig suggests.
The investigate was funded by the Wyss Institute the Paul G. Allen Frontiers Team the Harvard University Center for AIDS Research, which is supported by the Countrywide Institutes of Overall health a Burroughs-Wellcome American Modern society of Tropical Drugs and Cleanliness postdoctoral fellowship an American Gastroenterological Affiliation Takeda Pharmaceutical Investigation Scholar Award and an MIT-TATA Middle fellowship.




